Increasing data-efficiency in medical image analysis
Summary
Artificial intelligence has the opportunity to cause a huge disruption in the way healthcare operates, but gathering enough data for training remains a challenge. So what if, instead of focusing all our energy on this data collection process, we can adjust our deep learning algorithms to require less data?
To tackle this, we introduce Group Equivariant Convolutional Networks – networks with a convolutional layer that generalises the weight sharing property to other types of transformations.
Group-Equivariant Convolutional Neural Networks:
- Reduce the data requirements for medical image analysis by
10x - Invariant not only to translation, but also rotation and reflection
- Proved effective for pulmonary nodule detecion
- Far better than data augmentation
Deep learning, and convolutional neural networks in particular, have rapidly become the methodology of choice for all (medical) image related tasks. However, these techniques typically require a substantial amount of labeled data to learn from, meaning a human radiologist needs to manually record their findings in a way the computer can understand.
 Furthermore, if we want the algorithms to generalise well over different patient populations, scanner types, reconstruction techniques and so forth, we need even more data!
Furthermore, if we want the algorithms to generalise well over different patient populations, scanner types, reconstruction techniques and so forth, we need even more data!
This presents us with the challenge of
The Intuition
To explore how we can improve the data efficiency of convolutional neural networks (CNN), we first need to understand why CNNs are such an appropriate choice for image tasks in the first place. The great thing about CNNs are that they are roughly invariant to


This is great for images – after all, it rarely matters where exactly in an image a structure occurs, as long as you can see that it’s there. To get technical here for a moment, translations are a type of transformation you can apply to the image, but whether or not you apply it has no influence on the prediction of the model. However, the problem is that there are other types of transformations, such as




This is a problem, because this means that you can’t just present your algorithm with one orientation of an object (such as a dog), and expect it to work with a similar object, but rotated or flipped. In practice however, especially in the medical domain, the things you want to recognise and detect can occur in every orientation, both on a small and large scale. A good example of this are pulmonary nodules, but this also goes for other types of lesions. In order for the model to be able to recognise that, you have to present the algorithm with all these orientations separately while training, which – you guessed it – means you need more training data.
Our solution to this was to create a new type of convolutional neural network (CNN) called the
Applying to Real World Problems
But enough about dogs. What about real problems, like medical findings? As a case study, we use
Pulmonary nodules are small lesions in the lung that may be indicative of lung cancer, the leading cause of cancer-related deaths worldwide. Radiologists will generally try to detect nodules in an early stage, so they can track the growth over time. However, looking for these nodules can feel like looking for a needle in a haystack - without the advantage that you can just burn down the haystack to find said needle.
Pulmonary nodules (also often called
That’s where
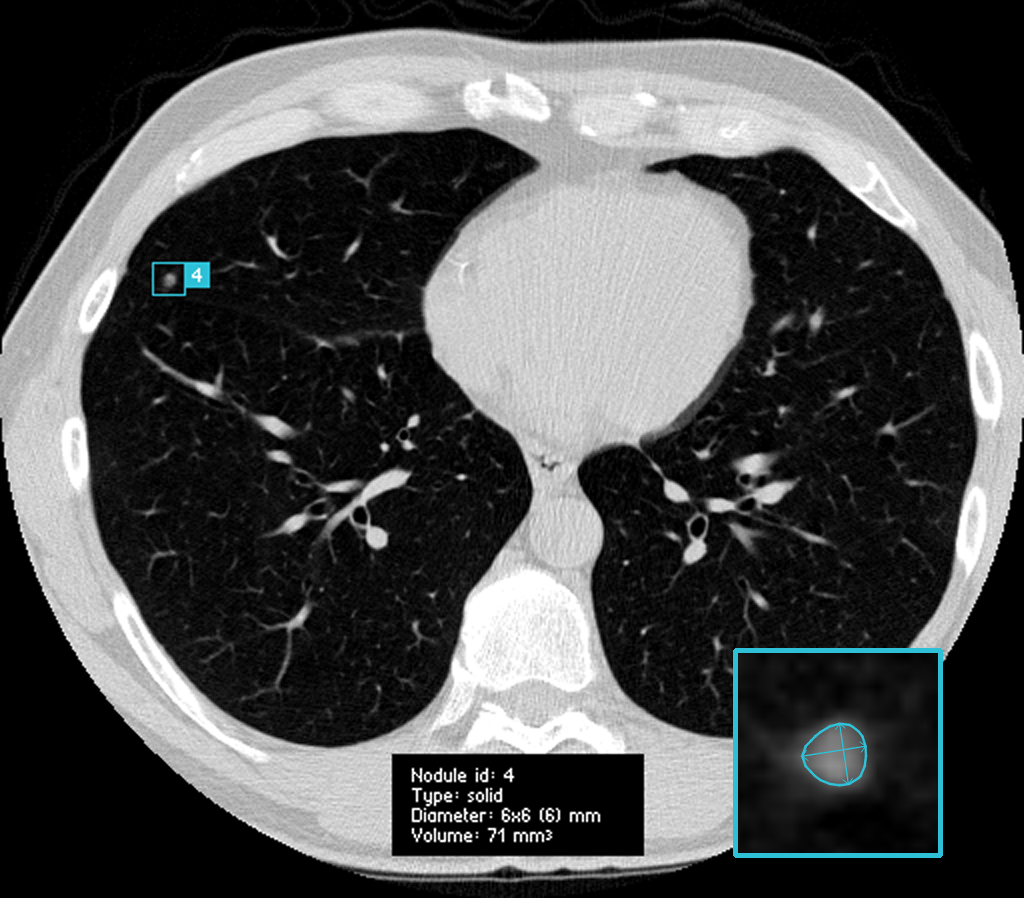
Normally, when approaching a data science problem, you simply utilise all the data you have available. Luckily, through the efforts of the
That’s why we trained our model on four different dataset sizes:
Did it work?
The results were astonishing. Of course, we hoped that our intuition was correct and we’d achieve an increase in performance, or a similar performance with a model trained on less data. What our experiments showed, however, was that the models trained on
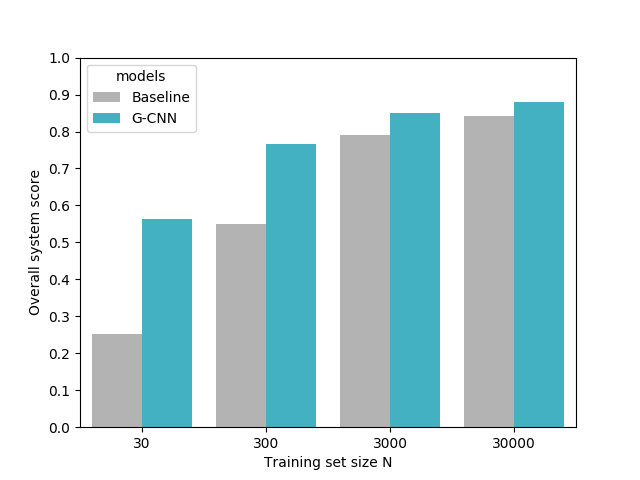
References
This research was performed as part of my thesis for the MSc Artificial Intelligence at the University of Amsterdam. It was supervised by Taco Cohen (Machine Learning researcher at Qualcomm) & prof. dr. Max Welling (research chair in Machine Learning at the University of Amsterdam and a VP Technologies at Qualcomm), as they originally laid the foundation of the work on equivariance and group-convolutional neural networks.
Implementations for 2D and 3D group-convolutions in Tensorflow, PyTorch and Chainer can be easily used from the GrouPy python package. A Keras implementation for 2D can be found here.
M. Winkels, T.S. Cohen, 3D G-CNNs for Pulmonary Nodule Detection, International Conference on Medical Imaging with Deep Learning (MIDL), 2018. [arxiv] [pdf]M. Winkels, T.S. Cohen, Pulmonary Nodule Detection in CT Scans with Equivariant CNNs, Under review at Medical Image Analysis (MIA), 2018. [pdf]
M. Winkels, T.S. Cohen, 3D Group-Equivariant Neural Networks for Octahedral and Square Prism Symmetry Groups, FAIM/ICML Workshop on Towards learning with limited labels: Equivariance, Invariance, and Beyond (ICML), 2018. [pdf]
T.S. Cohen, M. Welling, Group Equivariant Convolutional Networks, Proceedings of the International Conference on Machine Learning (ICML), 2016. [arxiv] [pdf]